Data Pipeline
Data flow schematic
Micromet Data Flow (currently for BB1, BB2, DSM, and RBM)
Note: Manitoba sites have SmartFlux stream only
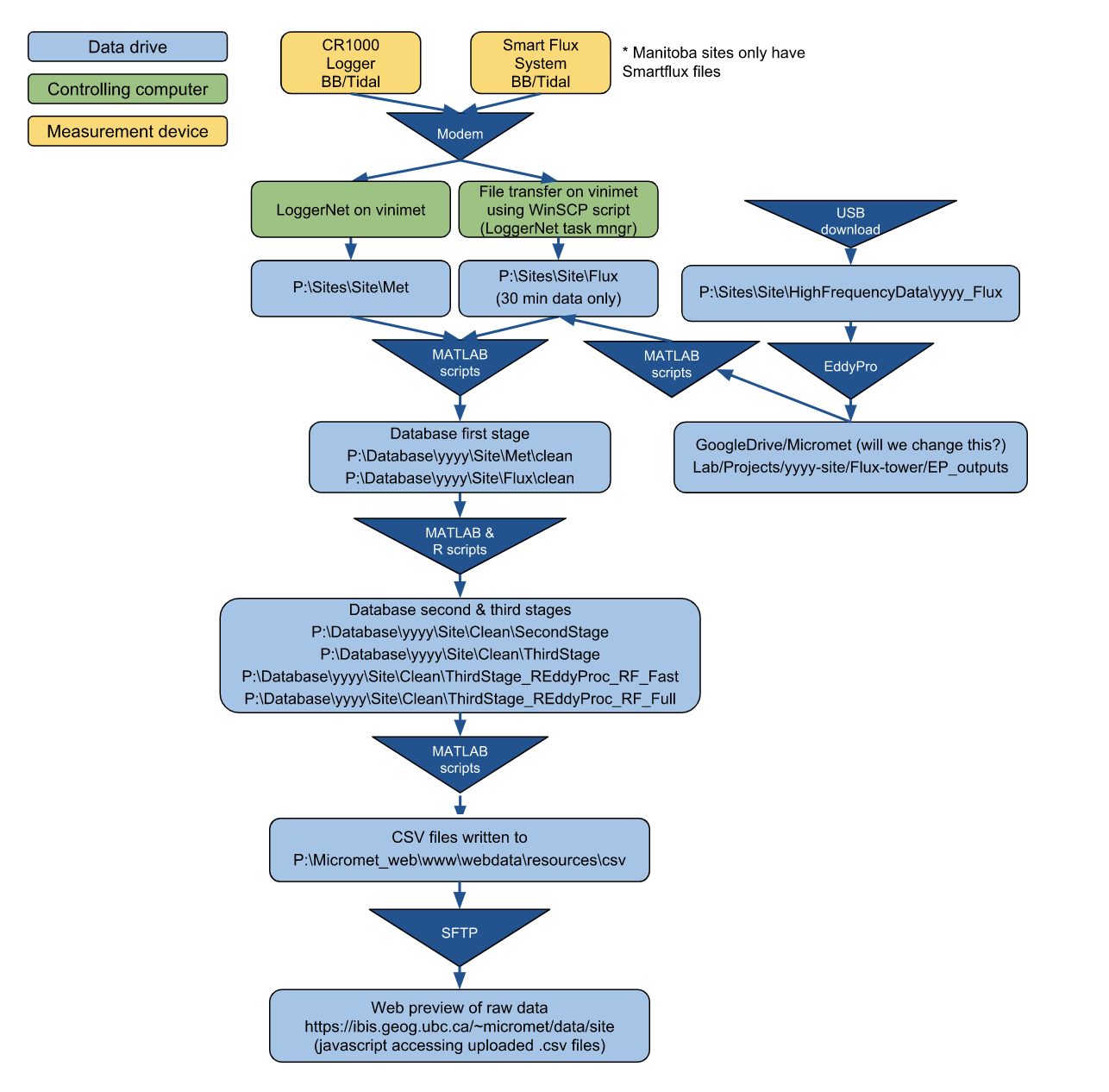
Drive/Folder structure on vinimet
P:\Sites\Site(data as received from the sites)\Met \5min (for BB only) \Flux \HighFrequencyDataP:\Database(everything in read_bor format)\yyyy\Sites \Met (straight from CRBasic -> read_bor) \Clean (First stage clean) \Flux (from EddyPro summary, site files -> read_bor & then overwrite with EddyPro recalculated output) \Clean (First stage clean) \Clean\SecondStage \ThirdStage \Ameriflux \ThirdStage_REddyProc_RF_Fast (simple uncertainty analysis) \ThirdStage_REddyProc_RF_Full (full uncertainty analysis)P:\Micromet_web(note that this mirrors what’s on host remote.geog.ubc.ca)\www \data (html & javascript for site level web plotting) \webdata\resources\csv (csv files for web plotting)Google drivecontains:- Pictures (to remove and host on google photos)
- Documents
Data stream notes
The data processing is run from LoggerNet Task manager through batch files stored under c:\UBC_Flux\BiometFTPsite. The batch files are named: Site_automatic_processing.bat. The generic Matlab scripts that’s called from these batch files is: run_BB_db_update([],{‘Site’},0)
More Details
MET Files
- Every 4 hrs (e.g., BB1 4:20, BB2 4:10) Loggernet Task Master downloads MET data to
P:\Sites\Site\Met\Site_MET.dat(or Site_biomet.dat)
Note Site is used generically to refer to each site (e.g., BB, BB2, DSM, RBM, Hogg, Young)
- Every 4 hrs (e.g., BB1 4:20, BB2 4:10) Loggernet Task Master downloads MET data to
Once per day 1:10 am
- ‘Rename files for all sites’ - Called from Loggernet Task Master
- Renames Site_MET.dat to Site_MET.yyyyddd
- -nosplash /r “renam_csi_dat_files”(
'P:Sites\Site\Met');…for each site; exit;”
- Site_Process_CSI_to_database - Called from Loggernet Task Master
-nosplash -minimize /r “run_BB_db_update([],{‘Site’});”
function run_BB_db_update(yearln) dv = datevec(now); arg_defualt('yearln', dv(1)); sites = {'Site'}; db_update_BB_site(yearln, sites); exit
- db_update_BB_site
- Update Site matlab database with new logger files to
P:\Database\yyyy\Site\Met - Call
C:\UBC_PC_Setup\PC_specific\BB_webupdateto create .csv files used by webplots -> `P:_web - Call
C:\Ubc_flux\BiometFTPsite\BB_Web_Update.batto move csv files to webserver
- Update Site matlab database with new logger files to
- ‘Rename files for all sites’ - Called from Loggernet Task Master
Location of data
P:- local drive on vinimetP:\Micromet_web\www\webdata\resources\csv- location of CSV filesP:\Micromet_web\www\data- local versions of js, html filesP:\Sites\Sites- raw data
Web-plotting pipeline
First, go from the bottom of the chart in the first page upwards, find out which step stopped working then use the followiwng instructions to troubleshoot.
EC Data
- SmartFlux files (*_EP-Summary.txt) arrive in the folders as below:
\\vinimet.geog.ubc.ca\Projects\Sites\BB\Flux\\vinimet.geog.ubc.ca\Projects\Sites\BB2\Flux
insert image
- Troubleshooting for Step 1:
If there is NO recent data files (.zip or .txt) at all, then:
- Check if you can still connect to LI-7200 to determine if it is an Internet issue or not.
- Check if the LoggerNet task for data downloads failed. When all tasks are executed properly, the status of the tasks should be shown like this:
insert image
- Check if the SmartFlux system at site skips processing or not by using WinSCP connecting to it (e.g., BB_site or BB2_site). The second figure below shows the BB2 SmartFlux have skipped processing of Feb. 1 (there is no summary file for that day):
insert images
If SmartFlux files arrive, the task “XX_automatic_processing” (as shown in the figure above) that runs Matlab program will process all data (CSI and SmartFlux) and update the web files too.
- Troubleshooting for Step 2:
- If you suspect that this task failed, then:
- Check the dates of the last updated web csv files (
\\VINIMET.GEOG.UBC.CA\Micromet_web\www\webdata\resources\csv). - Check a few flux-related csv files to see if the last datestamps are less than 6 hours old.
- If the files are NOT up-to-date, manually rerun the Matlab program by using
run_BB_db_update([]{'BB2},0)
- Check the dates of the last updated web csv files (
- If you confirm the task is working fine or you’ve re-run the Matlab program, then:
- Check if those files were uploaded to the web server by starting WinSCP to connect to ‘remote.geog.ubc.ca’ and see if the files are there.
- If you suspect that this task failed, then:
- Troubleshooting for Step 2:
Met Data
Data from dataloggers goes to \\vinimet.geog.ubc.ca\Projects\Sites\BB\Met\ or \\vinimet.geog.ubc.ca\Projects\Sites\BB2\Met\.
- Troubleshooting:
- Check if the files have arrived or not.
- Check the LoggerNet task for data downloads.
- Check connection to the logger
- Check the data of the last updates Biomet database files (
\\VINIMET.GEOG.UBC.CA\Database) - Check a few MET-related csv files to see if the last datestamps are less than 6 hours old.
- Manually re-run the task in Matlab by using
run_BB_db_update([]{'BB2},0). - Check if those csv files were uploaded to the web server.
Continue adding info from here
Other Documentation & Tasks
The following documents in the General procedures, settings and protocols folder in Google Drive may also be of use:
- Updating webplots →
Micromet web plotting.docx - Documents for dealing with full internal memory for SmartFlux →
Note_SmartFlux internal memory_ssh connection.docx & Note_SmartFlux internal memory_updater.docx - 7200 lab calibration →
Note_LI-7200 Lab calibration procedure.docx - Gas tank calibration →
Report_Gas tank calibration.docx
NOTE: Note that the troubleshooting reports folder contains documentation on troubleshooting that is relevant to all sites.
Creating a local copy of the database
To create a local copy of the Micromet Lab database or create your own database following the Micromet Lab database structure, following the instructions provided here